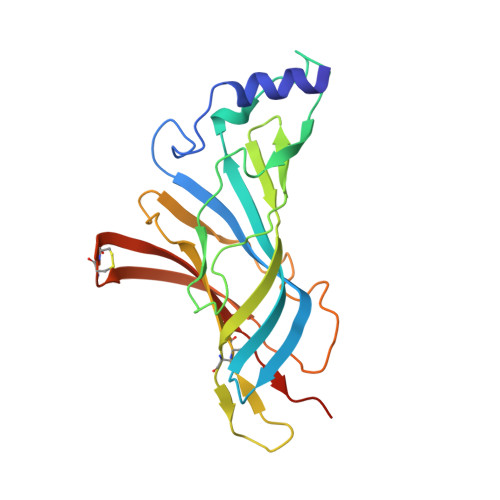
Crystal Structure of a Human Neuronal Nachr Extracellular Domain in Pentameric Assembly: Ligand-Bound Alpah2 Homopentamer.
Kouvatsos, N., Giastas, P., Chroni-Tzartou, D., Poulopoulou, C., Tzartos, S.J.(2016) Proc Natl Acad Sci U S A 113: 9635
- PubMed: 27493220
- DOI: https://doi.org/10.1073/pnas.1602619113
- Primary Citation of Related Structures:
5FJV - PubMed Abstract:
In this study we report the X-ray crystal structure of the extracellular domain (ECD) of the human neuronal α2 nicotinic acetylcholine receptor (nAChR) subunit in complex with the agonist epibatidine at 3.2 Å. Interestingly, α2 was crystallized as a pentamer, revealing the intersubunit interactions in a wild type neuronal nAChR ECD and the full ligand binding pocket conferred by two adjacent α subunits. The pentameric assembly presents the conserved structural scaffold observed in homologous proteins, as well as distinctive features, providing unique structural information of the binding site between principal and complementary faces. Structure-guided mutagenesis and electrophysiological data confirmed the presence of the α2(+)/α2(-) binding site on the heteromeric low sensitivity α2β2 nAChR and validated the functional importance of specific residues in α2 and β2 nAChR subunits. Given the pathological importance of the α2 nAChR subunit and the high sequence identity with α4 (78%) and other neuronal nAChR subunits, our findings offer valuable information for modeling several nAChRs and ultimately for structure-based design of subtype specific drugs against the nAChR associated diseases.
Organizational Affiliation:
Department of Neurobiology, Hellenic Pasteur Institute, 11521 Athens, Greece; nkouvatsos@pasteur.gr tzartos@pasteur.gr.